In this exercise we will learn how to do analyses using PGLS.
First, we will need a few libraries installed.
library(ape)
library(geiger)
library(nlme)
library(phytools)
## Loading required package: maps
## Loading required package: rgl
## Warning: failed to assign RegisteredNativeSymbol for getData to getData
## since getData is already defined in the 'phangorn' namespace
setwd("~/Documents/teaching/revellClass/2014bogota")
Second, we will need some data. We can read in anolis data and a phylogenetic tree. You can download the files from the following addresses:
anolisDataAppended.csv
anolis.phy
Download these files and place them in your working directory.
anoleData <- read.csv("anolisDataAppended.csv", row.names = 1)
anoleTree <- read.tree("anolis.phy")
Let’s see what this tree looks like.
plot(anoleTree)
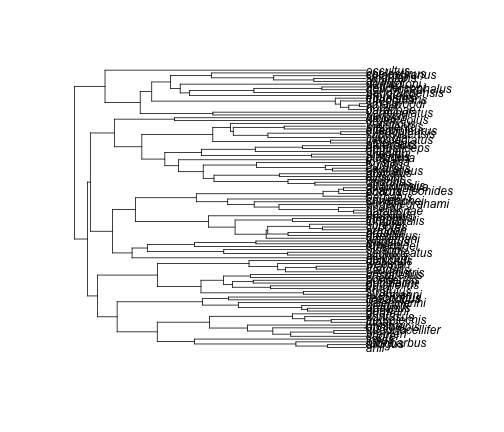
Geiger has a function to check that the names match between the tree and the data frame.
name.check(anoleTree, anoleData)
## [1] "OK"
Is there a correlation between awesomeness and hostility?
plot(anoleData[, c("awesomeness", "hostility")])
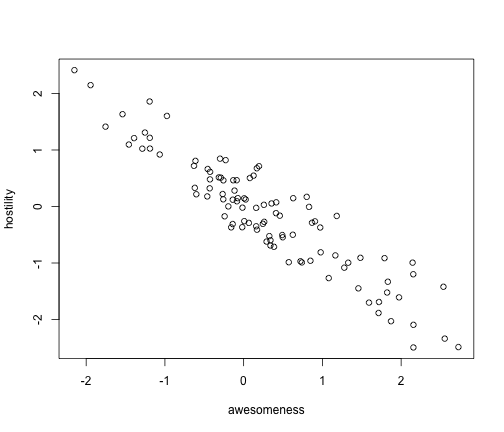
It certainly looks like there is. We can do this analysis easily with PICs, as you just learned:
# Extract columns
host <- anoleData[, "hostility"]
awe <- anoleData[, "awesomeness"]
# Give them names
names(host) <- names(awe) <- rownames(anoleData)
# Calculate PICs
hPic <- pic(host, anoleTree)
aPic <- pic(awe, anoleTree)
# Make a model
picModel <- lm(hPic ~ aPic - 1)
# Yes, significant
summary(picModel)
##
## Call:
## lm(formula = hPic ~ aPic - 1)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.105 -0.419 0.010 0.314 4.999
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## aPic -0.9776 0.0452 -21.6 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.897 on 98 degrees of freedom
## Multiple R-squared: 0.827, Adjusted R-squared: 0.825
## F-statistic: 469 on 1 and 98 DF, p-value: <2e-16
# plot results
plot(hPic ~ aPic)
abline(a = 0, b = coef(picModel))
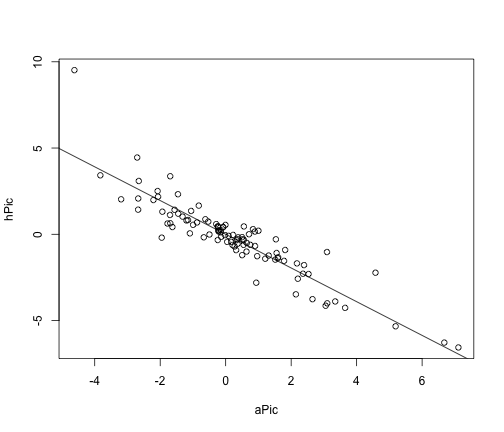
This whole procedure can be carried out more simply using PGLS.
pglsModel <- gls(hostility ~ awesomeness, correlation = corBrownian(phy = anoleTree),
data = anoleData, method = "ML")
summary(pglsModel)
## Generalized least squares fit by maximum likelihood
## Model: hostility ~ awesomeness
## Data: anoleData
## AIC BIC logLik
## 191 198.8 -92.49
##
## Correlation Structure: corBrownian
## Formula: ~1
## Parameter estimate(s):
## numeric(0)
##
## Coefficients:
## Value Std.Error t-value p-value
## (Intercept) 0.1506 0.26263 0.573 0.5678
## awesomeness -0.9776 0.04516 -21.648 0.0000
##
## Correlation:
## (Intr)
## awesomeness -0.042
##
## Standardized residuals:
## Min Q1 Med Q3 Max
## -0.76020 -0.39057 -0.04942 0.19597 1.07374
##
## Residual standard error: 0.8877
## Degrees of freedom: 100 total; 98 residual
coef(pglsModel)
## (Intercept) awesomeness
## 0.1506 -0.9776
plot(host ~ awe)
abline(a = coef(pglsModel)[1], b = coef(pglsModel)[2])
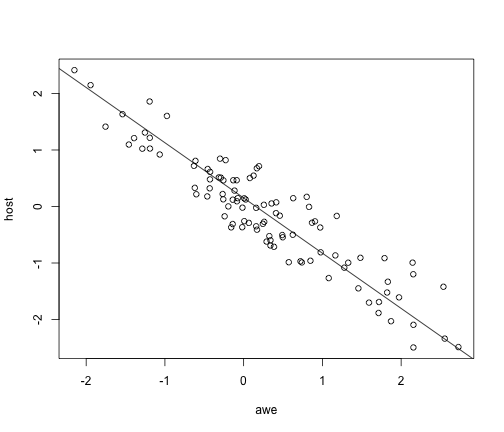
But PGLS is WAY more flexible than PICs. For example, we can include a discrete predictor:
pglsModel2 <- gls(hostility ~ ecomorph, correlation = corBrownian(phy = anoleTree),
data = anoleData, method = "ML")
anova(pglsModel2)
## Denom. DF: 93
## numDF F-value p-value
## (Intercept) 1 0.01847 0.8922
## ecomorph 6 0.21838 0.9700
coef(pglsModel2)
## (Intercept) ecomorphGB ecomorphT ecomorphTC ecomorphTG ecomorphTW
## 0.4844 -0.6316 -1.0585 -0.8558 -0.4086 -0.4039
## ecomorphU
## -0.7022
We can even include multiple predictors:
pglsModel3 <- gls(hostility ~ ecomorph * awesomeness, correlation = corBrownian(phy = anoleTree),
data = anoleData, method = "ML")
anova(pglsModel3)
## Denom. DF: 86
## numDF F-value p-value
## (Intercept) 1 0.1 0.7280
## ecomorph 6 1.4 0.2090
## awesomeness 1 472.9 <.0001
## ecomorph:awesomeness 6 3.9 0.0017
We can also assume that the error structure follows an OU model rather than Brownian motion:
# Does not converge - and this is difficult to fix!
pglsModelLambda <- gls(hostility ~ awesomeness, correlation = corPagel(1, phy = anoleTree,
fixed = FALSE), data = anoleData, method = "ML")
## Error: NA/NaN/Inf in foreign function call (arg 1)
# this is a problem with scale. We can do a quick fix by making the branch
# lengths longer. This will not affect the analysis other than rescaling a
# nuisance parameter
tempTree <- anoleTree
tempTree$edge.length <- tempTree$edge.length * 100
pglsModelLambda <- gls(hostility ~ awesomeness, correlation = corPagel(1, phy = tempTree,
fixed = FALSE), data = anoleData, method = "ML")
summary(pglsModelLambda)
## Generalized least squares fit by maximum likelihood
## Model: hostility ~ awesomeness
## Data: anoleData
## AIC BIC logLik
## 72.56 82.98 -32.28
##
## Correlation Structure: corPagel
## Formula: ~1
## Parameter estimate(s):
## lambda
## -0.1586
##
## Coefficients:
## Value Std.Error t-value p-value
## (Intercept) 0.0612 0.01582 3.872 2e-04
## awesomeness -0.8777 0.03104 -28.273 0e+00
##
## Correlation:
## (Intr)
## awesomeness -1
##
## Standardized residuals:
## Min Q1 Med Q3 Max
## -1.789463 -0.714775 0.003095 0.785093 2.232151
##
## Residual standard error: 0.371
## Degrees of freedom: 100 total; 98 residual
pglsModelOU <- gls(hostility ~ awesomeness, correlation = corMartins(1, phy = tempTree),
data = anoleData, method = "ML")
summary(pglsModelOU)
## Generalized least squares fit by maximum likelihood
## Model: hostility ~ awesomeness
## Data: anoleData
## AIC BIC logLik
## 96.63 107.1 -44.32
##
## Correlation Structure: corMartins
## Formula: ~1
## Parameter estimate(s):
## alpha
## 4.442
##
## Coefficients:
## Value Std.Error t-value p-value
## (Intercept) 0.1084 0.03953 2.743 0.0072
## awesomeness -0.8812 0.03658 -24.091 0.0000
##
## Correlation:
## (Intr)
## awesomeness -0.269
##
## Standardized residuals:
## Min Q1 Med Q3 Max
## -1.8665 -0.8133 -0.1104 0.6475 2.0919
##
## Residual standard error: 0.3769
## Degrees of freedom: 100 total; 98 residual